
July 14th 2020 - Press release
The results of the study, published in Nature Methods, may facilitate the development of new drugs related to diseases such as cancer and Alzheimer's. The new tool provides access to a large number of atomic-level simulations, the most basic, of the behaviour of a crucial element in almost all human physiological responses. This work is the result of a study by a consortium of international research centres led by scientists at the Hospital del Mar Medical Research Institute, in collaboration with Pompeu Fabra University and the Paul Scherrer Institute in Switzerland, and with the support of the Autonomous University of Barcelona.
G-protein coupled receptors (GPCRs) are one of the most basic elements in the response to stimuli such as a mosquito bite or the thrill of a goal scored by our favourite football team. In fact, these are the target of almost 40% of current drugs. But, until now, researchers had no tool that used molecular simulations to analyse and fully understand their function.

This is why a consortium of 23 institutions from 10 countries in Europe and the United States joined forces to design a tool to better understand the functioning of these receptors, whose job is to transmit signals inside the cell. The GPCRmd platform is the result of this work, led and coordinated by a group from the Hospital del Mar Medical Research Institute (IMIM) and Pompeu Fabra University's Biomedical Informatics Research Programme (GRIB) that looks at G-protein-coupled receptor-based drug development. Researchers from the Paul Scherrer Institute (PSI) in Switzerland and the Autonomous University of Barcelona also took part in the project.
Dr. Jana Selent, Miguel Servet researcher at the IMIM and the first author of the study, explains that "we used state-of-the-art technology, which requires very specific knowledge, including molecular simulations, to generate an online resource where scientists from different disciplines can easily inspect and analyse an exhaustive number of molecular simulations of GPCRs". This will allow them, according to Dr. Guixà-González, a postdoctoral researcher at the PSI in Switzerland and first co-author of the study, "to extract information relevant to their studies and develop new drugs related to diseases as serious as cancer and Alzheimer's". Other main authors are Ismael Rodríguez Espigares and Mariona Torrens Fontanals, postdoctoral and predoctoral researchers in the GRIB.
GPCRs are located in the cell membrane and transmit the information that triggers the body's physiological responses to the inside of the cell. For this reason, they are vitally important to many drugs. The new platform makes it possible to analyse their functioning on a very small scale, at the atomic level, with the help of newly developed software. Thanks to this, the simulations carried out by each of the groups that belong to the consortium, using a common protocol, were able to be transferred and visualised using a simple browser. This allows information to be exploited online and openly, for use by all researchers working in developing new treatments that target GPCRs.
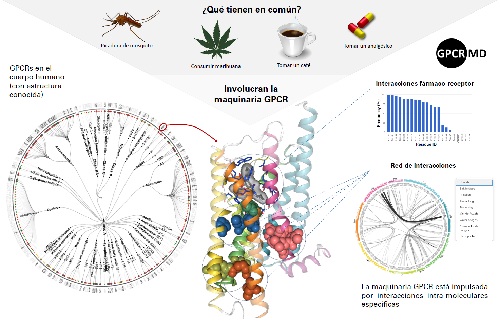
Currently, this tool covers approximately 70% of GPCRs with a known structure. The researchers aim to make this 100%, while at the same time they are creating a tool that will automatically perform the simulations and publish any new ones. They are not ruling out the possibility that, in the future, this system may be exported to other protein families of interest for the development of new drugs.
Rodriguez-Espigares et al. GPCRmd unveils the dynamics of the 3D-GPCRome. 2020, Nature Methods; https://www.nature.com/articles/s41592-020-0884-y


 This site complies with the HONcode standard for trustworthy health information:
This site complies with the HONcode standard for trustworthy health information: Parc Salut Mar
Passeig Marítim 25-29 Barcelona 08003
See location on Google maps
Phone: 93 248 30 00 · Fax: 93 248 32 54
Information request
© 2006 - 2024 Parc de Salut Mar · Legal notice and Privacy Police | Cookie Policy | Accessibility